
Research
Structural systems biology of microenvironmental stress and synthetic biology interventions
Abiotic factors such as extreme temperature, ionizing radiation, desiccation, and osmotic stress have complex consequences for biomolecules and cells, often challenging viability at multiple physical loci and affecting function of multiple processes. The Chang lab is focused on understanding the consequences of abiotic stresses spanning multiple scales of biological complexity from individual molecules to pathways and whole cells. Through integration of biological network modeling, protein structure analysis, machine learning, proteomics, and synthetic biology we seek to elucidate mechanisms of toxicity and resistance to microenvironmental stress and engineer translational protein interventions. Gaining better understanding and control over biological stability under abiotic stress has far-reaching applications, such as proteogenomic markers and improved therapies for cancer treatment, developing drought resistant crops, preservation of biologicals, engineering industrial and therapeutic microbes, and developing biotechnologies for long-term space travel.
Modeling and measuring microenvironmental stress phenotypes
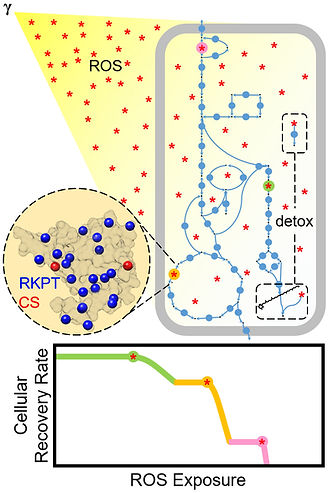
As a platform to study stress phenotypes, we develop network models to simulate cell growth and metabolism. These models can be integrated with structure analysis to predict viability-limiting proteins under stress conditions, such as associating bioinformatically-estimated protein melting temperatures with metabolic flux constraints to predict bottlenecks limiting growth under heat shock. A current goal of the lab in this area is to develop microbial and mammalian cell models to predict network-wide effects of oxidative stress caused by radiotherapy or chemotherapy. Experimentally-validated models could be used to identify prognostic markers of different healthy tissue and tumor types for guiding therapies, identify genetic targets to selectively sensitize tumors to radiotherapy, or design microbes that may serve as therapeutic probiotics to decrease toxicity of radiotherapy in sensitive healthy tissues such as the intestinal epithelium.
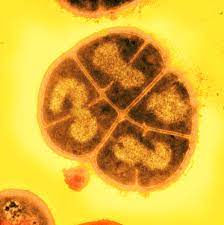
The polyextremophile Deinococcus radiodurans survives intense radiation in part due to protection of its proteome against oxidative damage, making it a valuable model organism for studying oxidative stress resistance.

Learning protein modifications from structure and proteomics
Advancing proteomic techniques have created an opportunity to study an important aspect of the protein structure-function relationship: post-translational modifications, both those serving native functions and those that consist of stress-induced damage. Our lab integrates proteomic techniques capable of detecting modified residues with proteome-wide structure analysis and machine learning techniques to uncover molecular properties that lead to these modifications. For example, this approach was used to identify predictive molecular properties of residues that get carbonylated under radiation-induced oxidative stress in E. coli and Deinococcus radiodurans. Having identified these predictive features, it is possible to extend the study of these modifications to sites and proteins that were not detected in proteomic experiments or to proteins from other organisms. These efforts enable us to differentiate sites and whole proteins that are more prone to modification or damage from those that are relatively more inert or robust, providing another angle by which to understand phenotypes in response to stress.
Protein engineering to protect against stress
Solutions to mitigate stress-induced damage at the molecular and cellular levels lie in synthetic biology. The residue-resolution understanding we are developing of what renders proteins sensitive to stress potentiates rational protein redesign to remove these vulnerabilities. We are currently developing such design approaches that could render essential targets that mediate cell death under oxidative stress more robust to damage and thereby also increase cell viability. Redesigned proteins can be analyzed by redox-proteomics to measure changes in sensitive sites and by activity assays in purified proteins and cell culture. In addition to the target-centric strategy to mitigate stress, our lab explores applications of intrinsically disordered proteins (IDPs) inspired by those from extremotolerant organisms, such as tardigrades and resurrection plants, to broadly protect biomolecules and whole cells from diverse environmental stresses. For example, expressing tardigrade damage suppressor protein (DSUP) in human cells increases their radioresistance by protecting against DNA damage.


Resurrection plants, like Selaginella lepidophylla, use IDPs to help them survive drought and rapidly recover upon rehydration. (time lapse by Shannon Nangle and Roger Chang)
